Decision Trees#
In this notebook you will experiment with different tree based algorithms on some different datasets. You will need to:
Create 3 models:
DecisionTreeClassifier()
RandomForestClassifier()
XGBClassifier()
These will be inserted into the dictionaries below for comparison.
You will also get a feeling for the different hyperparameters for each of these methods. Along with this, I want you to get used to reading the documentation of the different APIs. I will provide you with links.
https://scikit-learn.org/stable/modules/generated/sklearn.tree.DecisionTreeClassifier.html
https://scikit-learn.org/stable/modules/generated/sklearn.ensemble.RandomForestClassifier.html
https://xgboost.readthedocs.io/en/latest/python/python_api.html#module-xgboost.training
import numpy as np
import matplotlib.pyplot as plt
from sklearn.datasets import make_classification,make_moons
from sklearn.metrics import accuracy_score
from sklearn.model_selection import train_test_split
from matplotlib.colors import ListedColormap
from sklearn.datasets import make_circles
import time
from sklearn.tree import DecisionTreeClassifier
from sklearn.ensemble import RandomForestClassifier
from xgboost import XGBClassifier
/sciclone/home/jgiroux/.conda/envs/ai4fusion_2025/lib/python3.12/site-packages/xgboost/core.py:377: FutureWarning: Your system has an old version of glibc (< 2.28). We will stop supporting Linux distros with glibc older than 2.28 after **May 31, 2025**. Please upgrade to a recent Linux distro (with glibc >= 2.28) to use future versions of XGBoost.
Note: You have installed the 'manylinux2014' variant of XGBoost. Certain features such as GPU algorithms or federated learning are not available. To use these features, please upgrade to a recent Linux distro with glibc 2.28+, and install the 'manylinux_2_28' variant.
warnings.warn(
Example 1#
Instantiate your models using the default hyperparameters provided by the API.
Train them for both of the datasets provided below and note any differences you see. Note that in these simple datasets we will not be truly able to appreciate the power of the more advanced XGBoost.
# Create synthetic 2D dataset
X, y = make_circles(n_samples=1000, noise=0.2, factor=0.5, random_state=42)
# Train/test split
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.3, random_state=42)
# Classifiers
# Insert the 3 differnt models here under their associated names
# Hint: You will want to check what we have imported above and reference their class names in the above documentation
# For all of the models set the random state = 42
# In this first section you should expect
# Decision Tree - two arguments, max_depth=5 and random_state=42
# Random Forest - two arguments, n_estimators=100 and random_state=42
# XGBoost - two arguments, eval_metric="logloss" and random_state=42
models = {
"Decision Tree":
"Random Forest":
"XGBoost":
}
# Train and evaluate
results = {}
for name, model in models.items():
start = time.time()
model.fit(X_train, y_train)
train_time = time.time() - start
train_acc = accuracy_score(y_train, model.predict(X_train))
test_acc = accuracy_score(y_test, model.predict(X_test))
results[name] = (model, train_acc, test_acc, train_time)
print(f"{name}")
print(f" → Train Accuracy: {train_acc:.3f}")
print(f" → Test Accuracy: {test_acc:.3f}")
print(f" → Train Time : {train_time*1000:.2f} ms\n")
# Plot decision boundaries
def plot_decision_boundary(model, X, y, title):
h = 0.01
x_min, x_max = X[:, 0].min() - 1, X[:, 0].max() + 1
y_min, y_max = X[:, 1].min() - 1, X[:, 1].max() + 1
xx, yy = np.meshgrid(np.arange(x_min, x_max, h),
np.arange(y_min, y_max, h))
Z = model.predict(np.c_[xx.ravel(), yy.ravel()])
Z = Z.reshape(xx.shape)
cmap_light = ListedColormap(['#FFAAAA', '#AAAAFF'])
cmap_bold = ListedColormap(['#FF0000', '#0000FF'])
plt.figure(figsize=(6, 5))
plt.contourf(xx, yy, Z, cmap=cmap_light, alpha=0.6)
plt.scatter(X[:, 0], X[:, 1], c=y, cmap=cmap_bold, edgecolor='k')
plt.title(title)
plt.xlabel("Feature 1")
plt.ylabel("Feature 2")
plt.tight_layout()
plt.show()
# Visualize each model
for name, (model, train_acc, test_acc, _) in results.items():
plot_decision_boundary(model, X, y, f"{name}\nTrain Acc: {train_acc:.2f} | Test Acc: {test_acc:.2f}")
Another Dataset#
Copy your models code from above here.
# Create synthetic 2D dataset
X, y = make_moons(n_samples=1000, noise=0.3, random_state=42)
# Train/test split
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.3, random_state=42)
# Classifiers - copy from above
models = {
"Decision Tree":
"Random Forest":
"XGBoost":
}
# Train and evaluate
results = {}
for name, model in models.items():
start = time.time()
model.fit(X_train, y_train)
train_time = time.time() - start
train_acc = accuracy_score(y_train, model.predict(X_train))
test_acc = accuracy_score(y_test, model.predict(X_test))
results[name] = (model, train_acc, test_acc, train_time)
print(f"{name}")
print(f" → Train Accuracy: {train_acc:.3f}")
print(f" → Test Accuracy: {test_acc:.3f}")
print(f" → Train Time : {train_time*1000:.2f} ms\n")
# Plot decision boundaries
def plot_decision_boundary(model, X, y, title):
h = 0.01
x_min, x_max = X[:, 0].min() - 1, X[:, 0].max() + 1
y_min, y_max = X[:, 1].min() - 1, X[:, 1].max() + 1
xx, yy = np.meshgrid(np.arange(x_min, x_max, h),
np.arange(y_min, y_max, h))
Z = model.predict(np.c_[xx.ravel(), yy.ravel()])
Z = Z.reshape(xx.shape)
cmap_light = ListedColormap(['#FFAAAA', '#AAAAFF'])
cmap_bold = ListedColormap(['#FF0000', '#0000FF'])
plt.figure(figsize=(6, 5))
plt.contourf(xx, yy, Z, cmap=cmap_light, alpha=0.6)
plt.scatter(X[:, 0], X[:, 1], c=y, cmap=cmap_bold, edgecolor='k')
plt.title(title)
plt.xlabel("Feature 1")
plt.ylabel("Feature 2")
plt.tight_layout()
plt.show()
# Visualize each model
for name, (model, train_acc, test_acc, _) in results.items():
plot_decision_boundary(model, X, y, f"{name}\nTrain Acc: {train_acc:.2f} | Test Acc: {test_acc:.2f}")
Step 2#
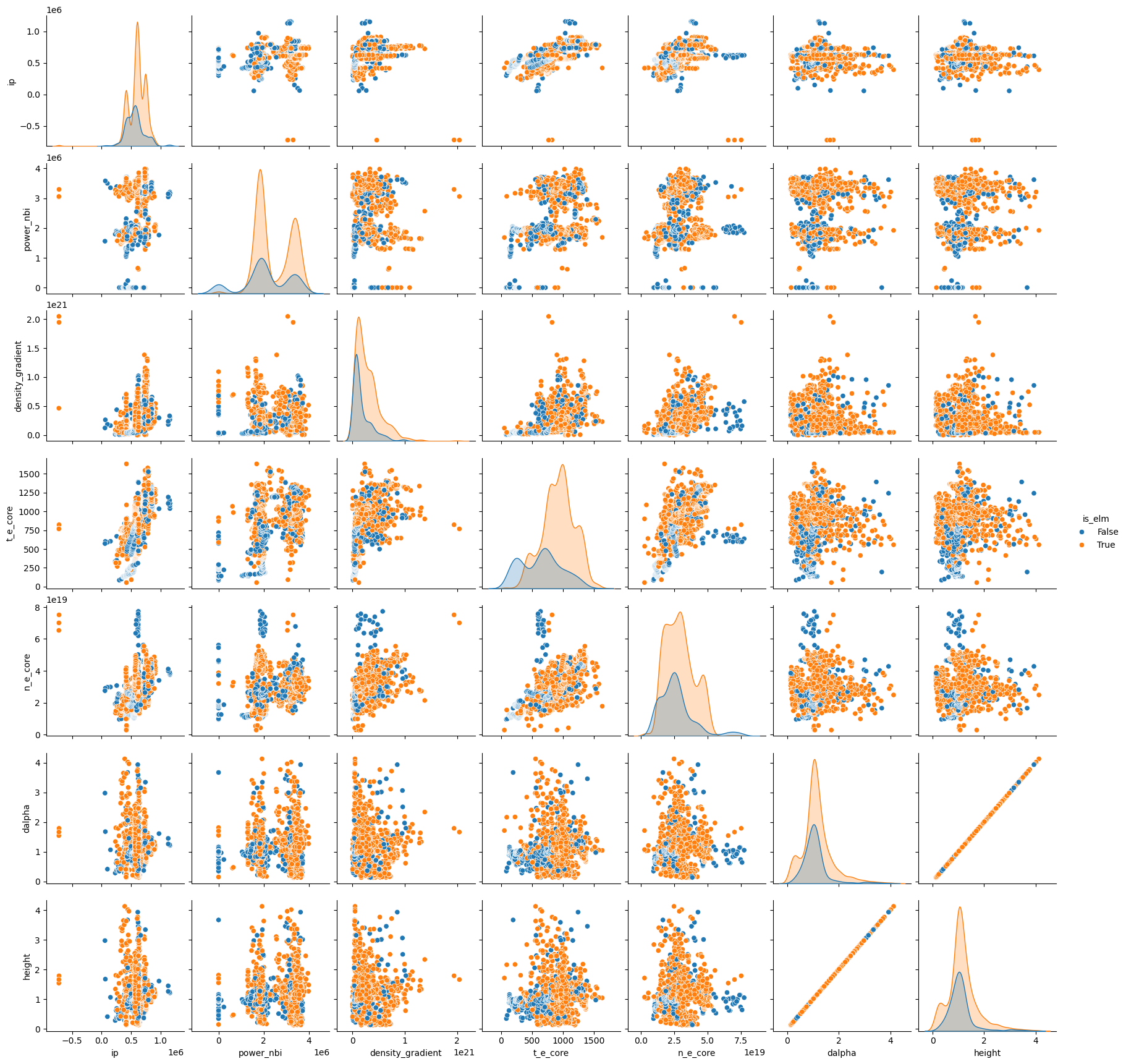
Play with the hyperparameter settings of each model and get a feel for what they do. You should use the ELM dataset and the same cross validation scheme created above. You can also change the number of folds used, along with the metric used for evaluation.
Things to look for:
Performance increases or decreases
Increased runtime
Overfitting
You can copy the code cells from above.
Step 3 (if time permits)#
Produce some feature importance plots from XGBoost. You may find plots of this nature useful for your final project.