Logistic Regression#
%pip install pytensor pymc
Requirement already satisfied: pytensor in /usr/local/lib/python3.10/dist-packages (2.18.6)
Requirement already satisfied: pymc in /usr/local/lib/python3.10/dist-packages (5.10.4)
Requirement already satisfied: setuptools>=48.0.0 in /usr/local/lib/python3.10/dist-packages (from pytensor) (67.7.2)
Requirement already satisfied: scipy>=0.14 in /usr/local/lib/python3.10/dist-packages (from pytensor) (1.11.4)
Requirement already satisfied: numpy>=1.17.0 in /usr/local/lib/python3.10/dist-packages (from pytensor) (1.25.2)
Requirement already satisfied: filelock in /usr/local/lib/python3.10/dist-packages (from pytensor) (3.14.0)
Requirement already satisfied: etuples in /usr/local/lib/python3.10/dist-packages (from pytensor) (0.3.9)
Requirement already satisfied: logical-unification in /usr/local/lib/python3.10/dist-packages (from pytensor) (0.4.6)
Requirement already satisfied: miniKanren in /usr/local/lib/python3.10/dist-packages (from pytensor) (1.0.3)
Requirement already satisfied: cons in /usr/local/lib/python3.10/dist-packages (from pytensor) (0.4.6)
Requirement already satisfied: typing-extensions in /usr/local/lib/python3.10/dist-packages (from pytensor) (4.12.1)
Requirement already satisfied: arviz>=0.13.0 in /usr/local/lib/python3.10/dist-packages (from pymc) (0.15.1)
Requirement already satisfied: cachetools>=4.2.1 in /usr/local/lib/python3.10/dist-packages (from pymc) (5.3.3)
Requirement already satisfied: cloudpickle in /usr/local/lib/python3.10/dist-packages (from pymc) (2.2.1)
Requirement already satisfied: fastprogress>=0.2.0 in /usr/local/lib/python3.10/dist-packages (from pymc) (1.0.3)
Requirement already satisfied: pandas>=0.24.0 in /usr/local/lib/python3.10/dist-packages (from pymc) (2.0.3)
Requirement already satisfied: matplotlib>=3.2 in /usr/local/lib/python3.10/dist-packages (from arviz>=0.13.0->pymc) (3.7.1)
Requirement already satisfied: packaging in /usr/local/lib/python3.10/dist-packages (from arviz>=0.13.0->pymc) (24.0)
Requirement already satisfied: xarray>=0.21.0 in /usr/local/lib/python3.10/dist-packages (from arviz>=0.13.0->pymc) (2023.7.0)
Requirement already satisfied: h5netcdf>=1.0.2 in /usr/local/lib/python3.10/dist-packages (from arviz>=0.13.0->pymc) (1.3.0)
Requirement already satisfied: xarray-einstats>=0.3 in /usr/local/lib/python3.10/dist-packages (from arviz>=0.13.0->pymc) (0.7.0)
Requirement already satisfied: python-dateutil>=2.8.2 in /usr/local/lib/python3.10/dist-packages (from pandas>=0.24.0->pymc) (2.8.2)
Requirement already satisfied: pytz>=2020.1 in /usr/local/lib/python3.10/dist-packages (from pandas>=0.24.0->pymc) (2023.4)
Requirement already satisfied: tzdata>=2022.1 in /usr/local/lib/python3.10/dist-packages (from pandas>=0.24.0->pymc) (2024.1)
Requirement already satisfied: toolz in /usr/local/lib/python3.10/dist-packages (from logical-unification->pytensor) (0.12.1)
Requirement already satisfied: multipledispatch in /usr/local/lib/python3.10/dist-packages (from logical-unification->pytensor) (1.0.0)
Requirement already satisfied: h5py in /usr/local/lib/python3.10/dist-packages (from h5netcdf>=1.0.2->arviz>=0.13.0->pymc) (3.9.0)
Requirement already satisfied: contourpy>=1.0.1 in /usr/local/lib/python3.10/dist-packages (from matplotlib>=3.2->arviz>=0.13.0->pymc) (1.2.1)
Requirement already satisfied: cycler>=0.10 in /usr/local/lib/python3.10/dist-packages (from matplotlib>=3.2->arviz>=0.13.0->pymc) (0.12.1)
Requirement already satisfied: fonttools>=4.22.0 in /usr/local/lib/python3.10/dist-packages (from matplotlib>=3.2->arviz>=0.13.0->pymc) (4.53.0)
Requirement already satisfied: kiwisolver>=1.0.1 in /usr/local/lib/python3.10/dist-packages (from matplotlib>=3.2->arviz>=0.13.0->pymc) (1.4.5)
Requirement already satisfied: pillow>=6.2.0 in /usr/local/lib/python3.10/dist-packages (from matplotlib>=3.2->arviz>=0.13.0->pymc) (9.4.0)
Requirement already satisfied: pyparsing>=2.3.1 in /usr/local/lib/python3.10/dist-packages (from matplotlib>=3.2->arviz>=0.13.0->pymc) (3.1.2)
Requirement already satisfied: six>=1.5 in /usr/local/lib/python3.10/dist-packages (from python-dateutil>=2.8.2->pandas>=0.24.0->pymc) (1.16.0)
import pymc as pm
import numpy as np
import pandas as pd
import seaborn as sns
import scipy.stats as stats
from scipy.special import expit as logistic
import matplotlib.pyplot as plt
import arviz as az
import requests
import io
az.style.use('arviz-darkgrid')
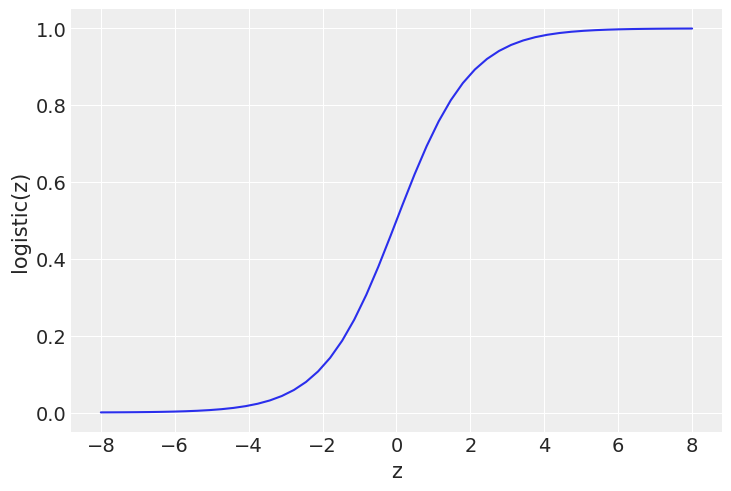
z = np.linspace(-8, 8)
plt.plot(z, 1 / (1 + np.exp(-z)))
plt.xlabel('z')
plt.ylabel('logistic(z)')
Text(0, 0.5, 'logistic(z)')
The Iris Dataset#
target_url = 'https://raw.githubusercontent.com/cfteach/brds/main/datasets/iris.csv'
download = requests.get(target_url).content
iris = pd.read_csv(io.StringIO(download.decode('utf-8')))
iris.head()
| sepal_length | sepal_width | petal_length | petal_width | species | |
|---|---|---|---|---|---|
| 0 | 5.1 | 3.5 | 1.4 | 0.2 | setosa |
| 1 | 4.9 | 3.0 | 1.4 | 0.2 | setosa |
| 2 | 4.7 | 3.2 | 1.3 | 0.2 | setosa |
| 3 | 4.6 | 3.1 | 1.5 | 0.2 | setosa |
| 4 | 5.0 | 3.6 | 1.4 | 0.2 | setosa |
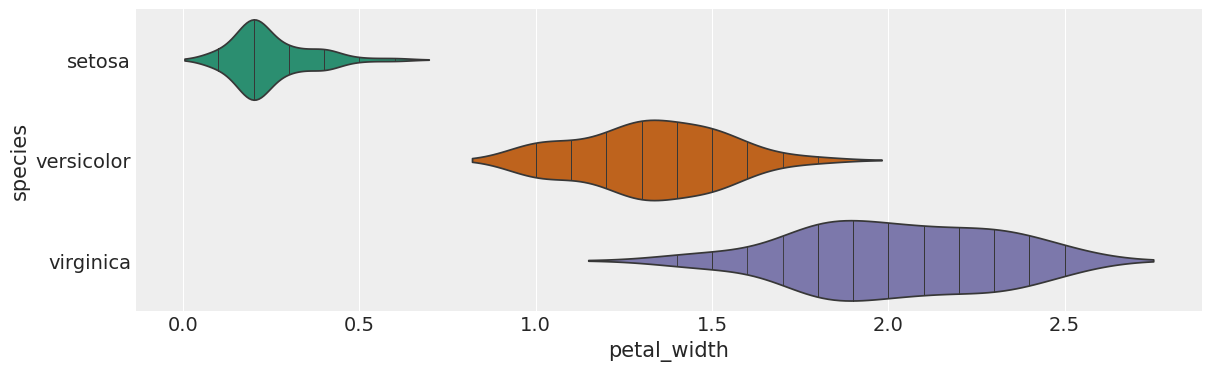
# species vs petal_width
from matplotlib import pyplot as plt
import seaborn as sns
figsize = (12, 1.2 * len(iris['species'].unique()))
plt.figure(figsize=figsize)
sns.violinplot(iris, x='petal_width', y='species', inner='stick', palette='Dark2')
sns.despine(top=True, right=True, bottom=True, left=True)
<ipython-input-9-caa5570effe5>:7: FutureWarning:
Passing `palette` without assigning `hue` is deprecated and will be removed in v0.14.0. Assign the `y` variable to `hue` and set `legend=False` for the same effect.
sns.violinplot(iris, x='petal_width', y='species', inner='stick', palette='Dark2')
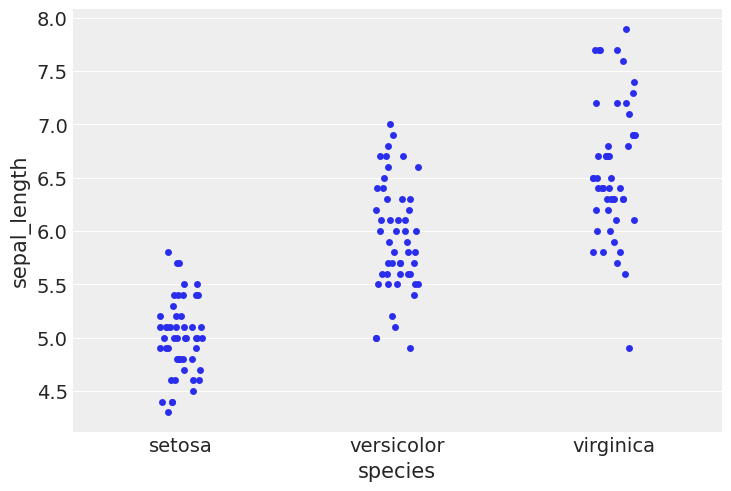
#using stripplot function from seaborn
sns.stripplot(x="species", y="sepal_length", data=iris, jitter=True)
<Axes: xlabel='species', ylabel='sepal_length'>
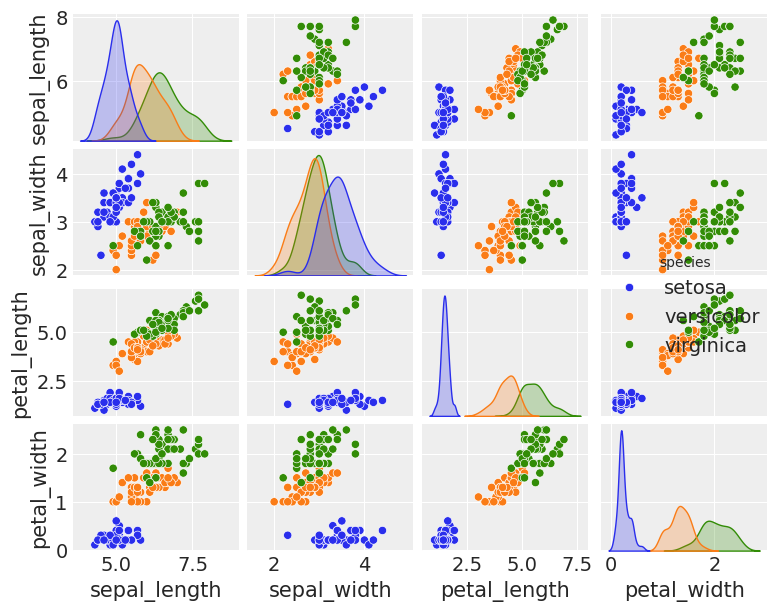
sns.pairplot(iris, hue='species', diag_kind='kde', height=1.5)
/usr/local/lib/python3.10/dist-packages/seaborn/axisgrid.py:123: UserWarning: The figure layout has changed to tight
self._figure.tight_layout(*args, **kwargs)
/usr/local/lib/python3.10/dist-packages/seaborn/axisgrid.py:213: UserWarning: This figure was using a layout engine that is incompatible with subplots_adjust and/or tight_layout; not calling subplots_adjust.
self._figure.subplots_adjust(right=right)
/usr/local/lib/python3.10/dist-packages/seaborn/axisgrid.py:123: UserWarning: The figure layout has changed to tight
self._figure.tight_layout(*args, **kwargs)
<seaborn.axisgrid.PairGrid at 0x7a327cda46a0>
The logistic model applied to the iris dataset#
df = iris.query("species == ('setosa', 'versicolor')")
df
| sepal_length | sepal_width | petal_length | petal_width | species | |
|---|---|---|---|---|---|
| 0 | 5.1 | 3.5 | 1.4 | 0.2 | setosa |
| 1 | 4.9 | 3.0 | 1.4 | 0.2 | setosa |
| 2 | 4.7 | 3.2 | 1.3 | 0.2 | setosa |
| 3 | 4.6 | 3.1 | 1.5 | 0.2 | setosa |
| 4 | 5.0 | 3.6 | 1.4 | 0.2 | setosa |
| ... | ... | ... | ... | ... | ... |
| 95 | 5.7 | 3.0 | 4.2 | 1.2 | versicolor |
| 96 | 5.7 | 2.9 | 4.2 | 1.3 | versicolor |
| 97 | 6.2 | 2.9 | 4.3 | 1.3 | versicolor |
| 98 | 5.1 | 2.5 | 3.0 | 1.1 | versicolor |
| 99 | 5.7 | 2.8 | 4.1 | 1.3 | versicolor |
100 rows × 5 columns
# converting the 'species' column of a DataFrame df into categorical codes using Pandas
y_0 = pd.Categorical(df['species']).codes
y_0
array([0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,
0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,
0, 0, 0, 0, 0, 0, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1,
1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1,
1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1], dtype=int8)
# let's select one variate
x_n = 'sepal_length'
x_0 = df[x_n].values
#print(x_0)
# let's center our dataset, as we have done in other exercises
x_c = x_0 - x_0.mean()
print(x_c)
[-0.371 -0.571 -0.771 -0.871 -0.471 -0.071 -0.871 -0.471 -1.071 -0.571
-0.071 -0.671 -0.671 -1.171 0.329 0.229 -0.071 -0.371 0.229 -0.371
-0.071 -0.371 -0.871 -0.371 -0.671 -0.471 -0.471 -0.271 -0.271 -0.771
-0.671 -0.071 -0.271 0.029 -0.571 -0.471 0.029 -0.571 -1.071 -0.371
-0.471 -0.971 -1.071 -0.471 -0.371 -0.671 -0.371 -0.871 -0.171 -0.471
1.529 0.929 1.429 0.029 1.029 0.229 0.829 -0.571 1.129 -0.271
-0.471 0.429 0.529 0.629 0.129 1.229 0.129 0.329 0.729 0.129
0.429 0.629 0.829 0.629 0.929 1.129 1.329 1.229 0.529 0.229
0.029 0.029 0.329 0.529 -0.071 0.529 1.229 0.829 0.129 0.029
0.029 0.629 0.329 -0.471 0.129 0.229 0.229 0.729 -0.371 0.229]
with pm.Model() as model_logreg:
α = pm.Normal('α', mu=0, sigma=10)
β = pm.Normal('β', mu=0, sigma=10)
#μ = α + pm.math.dot(x_c, β)
μ = α + β * x_c
θ = pm.Deterministic('θ', pm.math.sigmoid(μ))
bd = pm.Deterministic('bd', -α/β)
yl = pm.Bernoulli('yl', p=θ, observed=y_0)
idata_logreg = pm.sample(2000, tune = 2000, return_inferencedata=True)
100.00% [4000/4000 00:03<00:00 Sampling chain 0, 0 divergences]
100.00% [4000/4000 00:03<00:00 Sampling chain 1, 0 divergences]
varnames = ['α', 'β', 'bd']
res = az.summary(idata_logreg)
#print(res)
az.summary(idata_logreg)
/usr/local/lib/python3.10/dist-packages/arviz/utils.py:184: NumbaDeprecationWarning: The 'nopython' keyword argument was not supplied to the 'numba.jit' decorator. The implicit default value for this argument is currently False, but it will be changed to True in Numba 0.59.0. See https://numba.readthedocs.io/en/stable/reference/deprecation.html#deprecation-of-object-mode-fall-back-behaviour-when-using-jit for details.
numba_fn = numba.jit(**self.kwargs)(self.function)
| mean | sd | hdi_3% | hdi_97% | mcse_mean | mcse_sd | ess_bulk | ess_tail | r_hat | |
|---|---|---|---|---|---|---|---|---|---|
| α | 0.310 | 0.327 | -0.295 | 0.919 | 0.006 | 0.005 | 3204.0 | 2411.0 | 1.0 |
| β | 5.383 | 1.022 | 3.613 | 7.437 | 0.019 | 0.014 | 3075.0 | 2658.0 | 1.0 |
| θ[0] | 0.165 | 0.058 | 0.068 | 0.281 | 0.001 | 0.001 | 3771.0 | 2816.0 | 1.0 |
| θ[1] | 0.068 | 0.036 | 0.011 | 0.135 | 0.001 | 0.000 | 3734.0 | 2777.0 | 1.0 |
| θ[2] | 0.027 | 0.020 | 0.001 | 0.064 | 0.000 | 0.000 | 3677.0 | 2759.0 | 1.0 |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| θ[96] | 0.815 | 0.063 | 0.691 | 0.924 | 0.001 | 0.001 | 2902.0 | 2330.0 | 1.0 |
| θ[97] | 0.980 | 0.017 | 0.950 | 1.000 | 0.000 | 0.000 | 2851.0 | 2658.0 | 1.0 |
| θ[98] | 0.165 | 0.058 | 0.068 | 0.281 | 0.001 | 0.001 | 3771.0 | 2816.0 | 1.0 |
| θ[99] | 0.815 | 0.063 | 0.691 | 0.924 | 0.001 | 0.001 | 2902.0 | 2330.0 | 1.0 |
| bd | -0.057 | 0.060 | -0.171 | 0.057 | 0.001 | 0.001 | 3410.0 | 2825.0 | 1.0 |
103 rows × 9 columns
theta_post= idata_logreg.posterior['θ']
print(np.shape(theta_post))
(2, 2000, 100)
theta = idata_logreg.posterior['θ'].mean(axis=0).mean(axis=0)
idx = np.argsort(x_c)
np.random.seed(123)
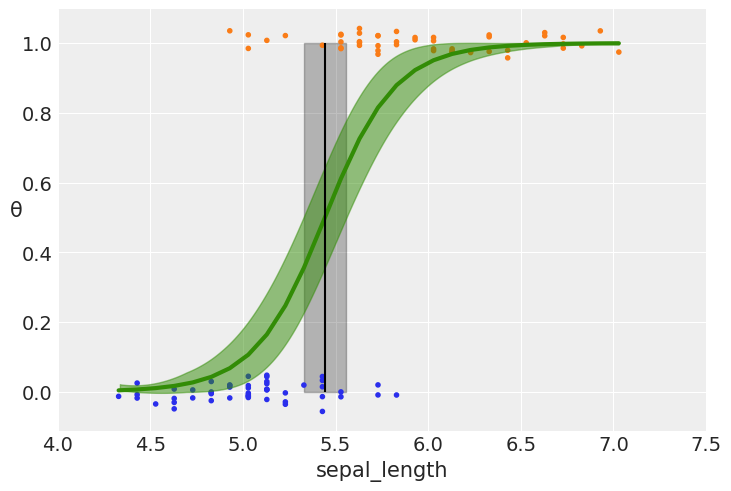
# plotting the sigmoid (logistic) curve
plt.plot(x_c[idx], theta[idx], color='C2', lw=3)
# plotting the mean boundary decision
plt.vlines(idata_logreg.posterior['bd'].mean(), 0, 1, color='k')
# ax.hdi will computer a lower and higher value of bd
bd_hdp = az.hdi(idata_logreg.posterior['bd'], hdi_prob=0.94)
# Fill the area between two vertical curves.
plt.fill_betweenx([0, 1], bd_hdp.bd[0].values, bd_hdp.bd[1].values, color='k', alpha=0.25)
plt.scatter(x_c, np.random.normal(y_0, 0.02),
marker='.', color=[f'C{x}' for x in y_0]) # 0.02 added for visualization purposes
az.plot_hdi(x_c, idata_logreg.posterior['θ'], color='C2') #green band
plt.xlabel(x_n)
plt.ylabel('θ', rotation=0)
# use original scale for xticks
locs, _ = plt.xticks()
plt.xticks(locs, np.round(locs + x_0.mean(), 1))
([<matplotlib.axis.XTick at 0x7a3276864550>,
<matplotlib.axis.XTick at 0x7a3276864820>,
<matplotlib.axis.XTick at 0x7a32769dde10>,
<matplotlib.axis.XTick at 0x7a3276b1ae00>,
<matplotlib.axis.XTick at 0x7a3276b189d0>,
<matplotlib.axis.XTick at 0x7a3276b19450>,
<matplotlib.axis.XTick at 0x7a3276b19f90>,
<matplotlib.axis.XTick at 0x7a3276b18c10>],
[Text(-1.5, 0, '4.0'),
Text(-1.0, 0, '4.5'),
Text(-0.5, 0, '5.0'),
Text(0.0, 0, '5.5'),
Text(0.5, 0, '6.0'),
Text(1.0, 0, '6.5'),
Text(1.5, 0, '7.0'),
Text(2.0, 0, '7.5')])
What happens if you change your priors?
Multidimensional (feature space) Logistic Regression#
df = iris.query("species == ('setosa', 'versicolor')")
y_1 = pd.Categorical(df['species']).codes
x_n = ['sepal_length', 'sepal_width']
x_1 = df[x_n].values
print(np.shape(x_1), type(x_1))
print(np.shape(y_1), type(y_1))
(100, 2) <class 'numpy.ndarray'>
(100,) <class 'numpy.ndarray'>
np.shape(x_1[:,0])
(100,)
with pm.Model() as model_1:
α = pm.Normal('α', mu=0, sigma=10)
β = pm.Normal('β', mu=0, sigma=2, shape=len(x_n))
#Data container that registers a data variable with the model.
#x__1 = pm.Data('x', x_1, mutable=True)
#y__1 = pm.Data('y', y_1, mutable=True)
# advantages: Model Reusability; Update Beliefs (reusing model); Efficeincy in sampling (same computational graph, even when data changes)
#Depending on the mutable setting (default: True), the variable is registered as a SharedVariable, enabling it to be altered in value and shape, but NOT in dimensionality using pymc.set_data().
μ = α + pm.math.dot(x_1, β)
θ = pm.Deterministic('θ', 1 / (1 + pm.math.exp(-μ)))
bd = pm.Deterministic('bd', -α/β[1] - β[0]/β[1] * x_1[:,0])
yl = pm.Bernoulli('yl', p=θ, observed=y_1)
trace_1 = pm.sample(1000, tune=2000, return_inferencedata=True, target_accept=0.9)
100.00% [3000/3000 00:29<00:00 Sampling chain 0, 0 divergences]
100.00% [3000/3000 00:31<00:00 Sampling chain 1, 0 divergences]
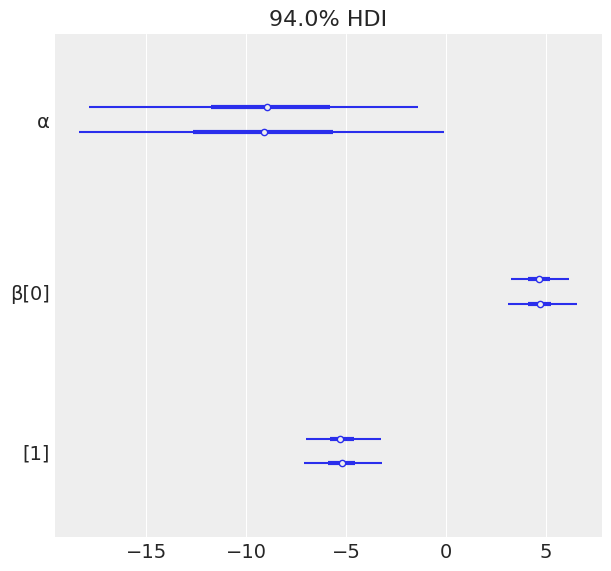
varnames = ['α', 'β']
az.plot_forest(trace_1, var_names=varnames);
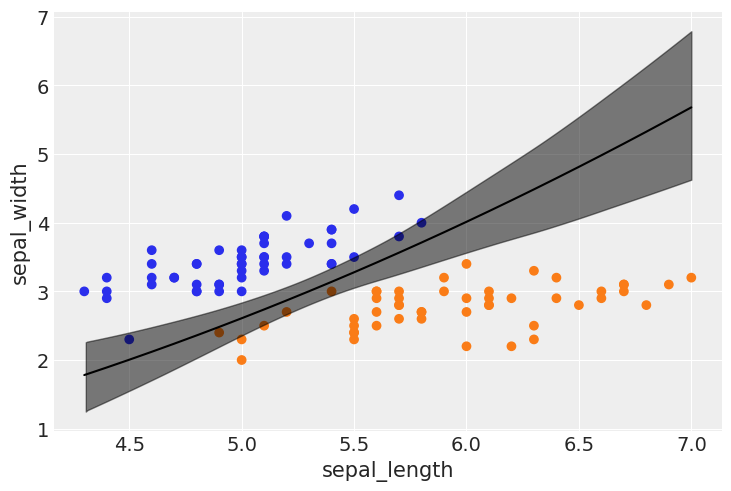
idx = np.argsort(x_1[:,0])
bd_mean = trace_1.posterior['bd'].mean(axis=0).mean(axis=0)
plt.scatter(x_1[:,0], x_1[:,1], c=[f'C{x}' for x in y_0])
bd = bd_mean[idx]
plt.plot(x_1[:,0][idx], bd, color='k');
az.plot_hdi(x_1[:,0], trace_1.posterior['bd'], color='k')
plt.xlabel(x_n[0])
plt.ylabel(x_n[1])
Text(0, 0.5, 'sepal_width')
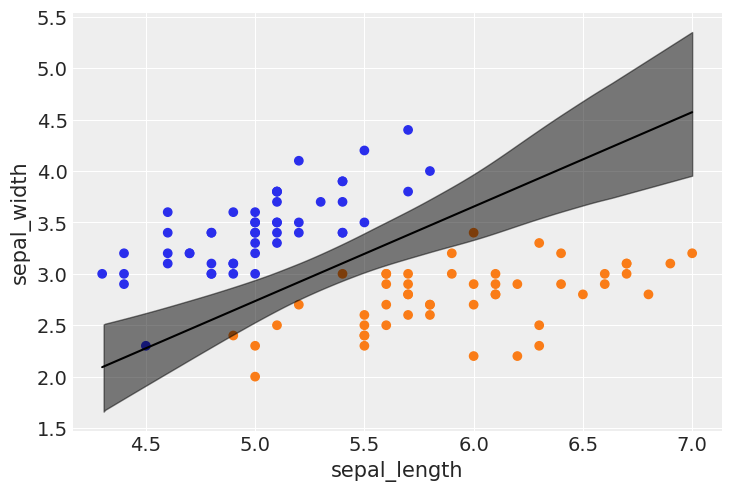
What happens if you change your priors?
What happens if your decision boundary is not linear? (See below)
Prediction on unseen data#
print(np.shape(x_1), type(x_1))
udata = np.array(((5.2,2.0),(7.0,2.0),(4.5,5.0),(5.5,3.2)))
print(np.shape(udata), type(udata))
(100, 2) <class 'numpy.ndarray'>
(4, 2) <class 'numpy.ndarray'>
print(udata)
[[5.2 2. ]
[7. 2. ]
[4.5 5. ]
[5.5 3.2]]
alpha_chain = trace_1.posterior['α'].mean(axis=0).values
beta_chain = trace_1.posterior['β'].mean(axis=0).values
print(np.shape(alpha_chain), np.shape(beta_chain))
(1000,) (1000, 2)
logit = np.dot(udata, beta_chain.T) + alpha_chain
print(np.shape(logit))
(4, 1000)
probabilities = 1 / (1 + np.exp(-logit))
print(np.shape(probabilities))
(4, 1000)
# Average probabilities for prediction
mean_probabilities = np.mean(probabilities, axis=1)
# Class assignment (you might adjust the threshold if needed, default is 0.5)
class_assignments = (mean_probabilities > 0.5).astype(int)
# Uncertainty estimation
lower_bound = np.percentile(probabilities, 2.5, axis=1)
upper_bound = np.percentile(probabilities, 97.5, axis=1)
print("\n=======================================")
print("data: \n", udata.T)
print("class, probabilities, ranges(94%HDI): ")
for h,i,j,k in zip(class_assignments, mean_probabilities, lower_bound,upper_bound):
print(f"class: {h}, mean prob. {i:.4f}, 94% HDI: [{j:.4f},{k:.4f}]")
print("=======================================\n")
=======================================
data:
[[5.2 7. 4.5 5.5]
[2. 2. 5. 3.2]]
class, probabilities, ranges(94%HDI):
class: 1, mean prob. 0.9898, 94% HDI: [0.9666,0.9986]
class: 1, mean prob. 1.0000, 94% HDI: [1.0000,1.0000]
class: 0, mean prob. 0.0000, 94% HDI: [0.0000,0.0000]
class: 0, mean prob. 0.4874, 94% HDI: [0.3275,0.6525]
=======================================
bd_mean = trace_1.posterior['bd'].mean(axis=0).mean(axis=0)
plt.scatter(udata[:,0], udata[:,1], c=[f'C{x}' for x in class_assignments])
bd = bd_mean[idx]
plt.plot(x_1[:,0][idx], bd, color='k');
az.plot_hdi(x_1[:,0], trace_1.posterior['bd'], color='k')
plt.xlabel(x_n[0])
plt.ylabel(x_n[1])
Text(0, 0.5, 'sepal_width')
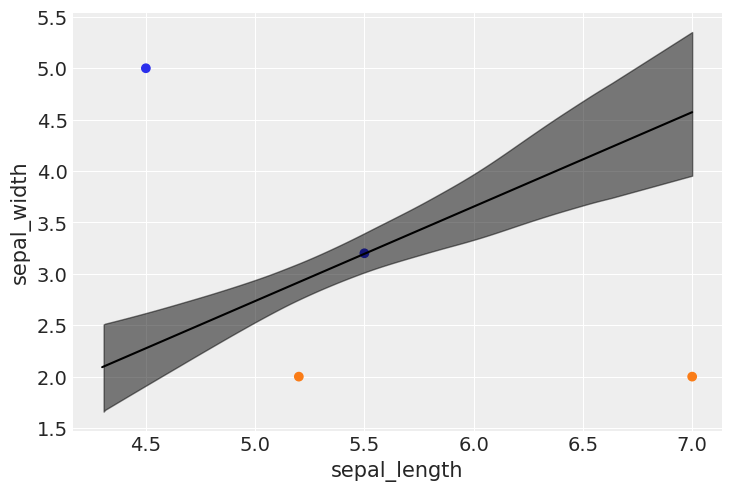
np.shape(x_1**2)
(100, 2)
x_1sq = x_1**2
print(np.shape(x_1sq), np.shape(x_1))
(100, 2) (100, 2)
Can I use a non-linear decision boundary?#
Let’s add, for example, a term \(\gamma \cdot x_{sepal \ length}^2\) in the equation for \(\mu\)
with pm.Model() as model_2:
α = pm.Normal('α', mu=0, sigma=10)
β = pm.Normal('β', mu=0, sigma=2, shape=len(x_n))
𝛾 = pm.Normal('𝛾', mu=0, sigma=2)
μ = α + pm.math.dot(x_1, β) + pm.math.dot(x_1[:,0]**2,𝛾)
θ = pm.Deterministic('θ', 1 / (1 + pm.math.exp(-μ)))
bd = pm.Deterministic('bd', -α/β[1] - β[0]/β[1] * x_1[:,0] - 𝛾/β[1] * x_1[:,0]**2)
yl = pm.Bernoulli('yl', p=θ, observed=y_1)
trace_2 = pm.sample(1000, tune=2000, return_inferencedata=True, target_accept=0.95)
100.00% [3000/3000 01:00<00:00 Sampling chain 0, 0 divergences]
100.00% [3000/3000 00:58<00:00 Sampling chain 1, 0 divergences]
trace_2
arviz.InferenceData
-
<xarray.Dataset> Dimensions: (chain: 2, draw: 1000, β_dim_0: 2, θ_dim_0: 100, bd_dim_0: 100) Coordinates: * chain (chain) int64 0 1 * draw (draw) int64 0 1 2 3 4 5 6 7 8 ... 992 993 994 995 996 997 998 999 * β_dim_0 (β_dim_0) int64 0 1 * θ_dim_0 (θ_dim_0) int64 0 1 2 3 4 5 6 7 8 9 ... 91 92 93 94 95 96 97 98 99 * bd_dim_0 (bd_dim_0) int64 0 1 2 3 4 5 6 7 8 ... 91 92 93 94 95 96 97 98 99 Data variables: α (chain, draw) float64 1.62 -7.419 0.1927 ... 11.85 13.82 13.3 β (chain, draw, β_dim_0) float64 -2.621 -7.707 ... -1.832 -5.801 𝛾 (chain, draw) float64 1.246 1.305 0.4872 ... 0.523 0.4905 0.4845 θ (chain, draw, θ_dim_0) float64 0.001814 0.01183 ... 0.8868 0.9136 bd (chain, draw, bd_dim_0) float64 2.681 2.426 2.183 ... 2.855 3.206 Attributes: created_at: 2024-02-22T19:31:16.798921 arviz_version: 0.15.1 inference_library: pymc inference_library_version: 5.7.2 sampling_time: 119.82878065109253 tuning_steps: 2000 -
<xarray.Dataset> Dimensions: (chain: 2, draw: 1000) Coordinates: * chain (chain) int64 0 1 * draw (draw) int64 0 1 2 3 4 5 ... 994 995 996 997 998 999 Data variables: (12/17) max_energy_error (chain, draw) float64 -0.2017 -0.03366 ... -0.3917 energy_error (chain, draw) float64 -0.02009 -0.008556 ... -0.2159 step_size (chain, draw) float64 0.04268 0.04268 ... 0.02932 perf_counter_start (chain, draw) float64 4.682e+03 ... 4.761e+03 acceptance_rate (chain, draw) float64 0.9935 0.9999 ... 0.8849 0.9788 energy (chain, draw) float64 20.74 21.26 ... 27.17 25.83 ... ... step_size_bar (chain, draw) float64 0.0381 0.0381 ... 0.03881 tree_depth (chain, draw) int64 7 6 7 7 7 7 7 5 ... 7 7 5 6 4 5 5 largest_eigval (chain, draw) float64 nan nan nan nan ... nan nan nan perf_counter_diff (chain, draw) float64 0.02136 0.01663 ... 0.005639 smallest_eigval (chain, draw) float64 nan nan nan nan ... nan nan nan reached_max_treedepth (chain, draw) bool False False False ... False False Attributes: created_at: 2024-02-22T19:31:16.812696 arviz_version: 0.15.1 inference_library: pymc inference_library_version: 5.7.2 sampling_time: 119.82878065109253 tuning_steps: 2000 -
<xarray.Dataset> Dimensions: (yl_dim_0: 100) Coordinates: * yl_dim_0 (yl_dim_0) int64 0 1 2 3 4 5 6 7 8 ... 91 92 93 94 95 96 97 98 99 Data variables: yl (yl_dim_0) int64 0 0 0 0 0 0 0 0 0 0 0 0 ... 1 1 1 1 1 1 1 1 1 1 1 Attributes: created_at: 2024-02-22T19:31:16.821619 arviz_version: 0.15.1 inference_library: pymc inference_library_version: 5.7.2
az.summary(trace_2)
| mean | sd | hdi_3% | hdi_97% | mcse_mean | mcse_sd | ess_bulk | ess_tail | r_hat | |
|---|---|---|---|---|---|---|---|---|---|
| α | -2.588 | 5.816 | -14.190 | 7.282 | 0.202 | 0.143 | 830.0 | 912.0 | 1.0 |
| β[0] | -0.215 | 1.781 | -3.216 | 3.336 | 0.067 | 0.051 | 704.0 | 624.0 | 1.0 |
| β[1] | -5.914 | 1.136 | -8.021 | -3.783 | 0.040 | 0.030 | 872.0 | 836.0 | 1.0 |
| 𝛾 | 0.765 | 0.261 | 0.259 | 1.238 | 0.009 | 0.007 | 786.0 | 930.0 | 1.0 |
| θ[0] | 0.016 | 0.014 | 0.000 | 0.043 | 0.000 | 0.000 | 925.0 | 985.0 | 1.0 |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| bd[95] | 3.564 | 0.157 | 3.255 | 3.842 | 0.004 | 0.003 | 1613.0 | 1583.0 | 1.0 |
| bd[96] | 3.564 | 0.157 | 3.255 | 3.842 | 0.004 | 0.003 | 1613.0 | 1583.0 | 1.0 |
| bd[97] | 4.326 | 0.301 | 3.756 | 4.865 | 0.008 | 0.006 | 1452.0 | 1530.0 | 1.0 |
| bd[98] | 2.737 | 0.117 | 2.510 | 2.941 | 0.003 | 0.002 | 1662.0 | 1525.0 | 1.0 |
| bd[99] | 3.564 | 0.157 | 3.255 | 3.842 | 0.004 | 0.003 | 1613.0 | 1583.0 | 1.0 |
204 rows × 9 columns
idx = np.argsort(x_1[:,0])
bd_mean2 = trace_2.posterior['bd'].mean(axis=0).mean(axis=0)
plt.scatter(x_1[:,0], x_1[:,1], c=[f'C{x}' for x in y_0])
bd = bd_mean2[idx]
plt.plot(x_1[:,0][idx], bd, color='k');
az.plot_hdi(x_1[:,0], trace_2.posterior['bd'], color='k')
plt.xlabel(x_n[0])
plt.ylabel(x_n[1])
print(np.shape(bd_mean2))
(100,)