Nonparamatric Methods #
The Nadaraya-Watson Regressor #
Main Idea: Estimate a conditional mean (expected value) given the input variables.
If we have estimates for the joint probability density \(f\) and the marginal density \(f_X\) we consider:
where \(X\) is the input data (features) and \(Y\) is the dependent variable.
We approximate the joint probability density function by using a kernel:
The marginal density is also approximated in a similar way:
By substituting these approximations into equation (1) we obtain:
Thus the conditional mean \(\mathbb{E}[Y| X=x]\) can be seen as a weighted average of \(y_i\):
where
This is to say that the Nadaraya-Watson estimator is a local mean of \(Y\) around \(X=x_i.\)
Functional Data Analysis - GAMs #
The main idea of generalized additive models (GAMs) is based on the assumption that
By comparison, a linear model is
We consider a regularized maximum likelihood approach for the generalized additive model
and for standardization we assume
For classificaiton problems we can use:
where \(g\) is a link function, e.g. the inverse of the logistic sigmoid.
To prevent overfitting, we consider a sparsity-smoothness regularization:
where the following term is designed to control the rapid accelerations/decelerations.
The objective function is
and this is to minimize over a suitable class of functions \(\mathcal{F}.\)
Cubic Splines#
What we need:
we need the tools, the building blocks for constructing any nonlinearity we want.
we need a good validation mechanism.
we need an efficient implementation of such tools in programming languages, e.g. Python.
The main idea is to use third degree polynomial functions, such as
for \(i=1,2,...,n-1\) and \(x_i\) are data points of the same feature. It is quite obvious that we have:
and $\( s''_i(x)=6a_i(x-x_i)+2b_i \)$
So we create
We generally want the following properties to hold:
\(S(x)\) is continuous on \([x_1,x_n].\)
\(S'(x)\) is continuous on \([x_1,x_n].\)
\(S''(x)\) is continuous on \([x_1,x_n].\)
And of course, we want to interpolate some data points:
and that is to say that
In order to make the curve smooth across the interval, the derivatives must be equal at the stitching points:
Let \(M_i:=2b_i\) and \(h=x_{i+1}-x_i\). After some sensible calculations we get the following matrix equation:
\begin{align} \begin{bmatrix} 1 & 4 & 1 & 0 & \dots & 0 & 0 & 0 & 0\ 0 & 1 & 4 & 1 & \dots & 0 & 0 & 0 & 0\ 0 & 0 & 1 & 4 & \dots & 0 & 0 & 0 & 0\ \vdots & \vdots & \vdots & \vdots & \ddots & \vdots & \vdots & \vdots & \vdots\ 0 & 0 & 0 & 0 & \dots & 4 & 1 & 0 & 0\ 0 & 0 & 0 & 0 & \dots & 1 & 1 & 1 & 0\ 0 & 0 & 0 & 0 & \dots & 0 & 1 & 4 & 1 \end{bmatrix} \begin{bmatrix} M_1\ M_2\ M_3\ M_4\ \vdots\ M_{n-3}\ M_{n-2}\ M_{n-1}\ M_n \end{bmatrix} =\frac{6}{h^2} \begin{bmatrix} y_1-2y_2+y_3\ y_2-2y_3+y_4\ y_3-2y_4+y_5\ \vdots\ y_{n-4}-2y_{n-3}+y_{n-2}\ y_{n-3}-2y_{n-2}+y_{n-1}\ y_{n-2}-2y_{n-1}+y_{n} \end{bmatrix} \end{align}
which linear system has \(n-2\) rows and \(n\) columns, so it is underdetermined. We can solve this system by imposing some extra conditions.
One easy condition is \(M_1=M_n=0\) and what we get is referred to as Natural Splines.
Generalized Additive Modeling#
The assumption is that
and of course, ideally, the mean of the noise should be 0; here \(p\) is the number of features.
So we get
This is the reson we need very good methods of regularization and variable selection.
Model Complexity#
Let’s assume that we have N knots. For each feature we fit N cubic splines. This means that we subdivide the range of each feature into N eaqually spaced intervals. On each such interval you have a cubic polynomial. For the same feature the cubic polynomials connect smoothly at the knots.
Model Complexity: we determine 4 coefficients for each cubic. We have N knots for each feature so in the end we have 4Np coefficients (weights) to determine.
Number of Splines If the data has \(n\) observations, we may consider
Code Applications#
Setup#
import os
if 'google.colab' in str(get_ipython()):
print('Running on CoLab')
from google.colab import drive
drive.mount('/content/drive')
os.chdir('/content/drive/My Drive/Data Sets')
!pip install -q pygam
else:
print('Running locally')
os.chdir('../Data')
Running on CoLab
Drive already mounted at /content/drive; to attempt to forcibly remount, call drive.mount("/content/drive", force_remount=True).
# library imports
import numpy as np
import pandas as pd
from math import ceil
from scipy import linalg
from sklearn.linear_model import LinearRegression, Ridge, Lasso, ElasticNet
from sklearn.metrics import mean_absolute_error, mean_squared_error as mse, r2_score
from sklearn.preprocessing import StandardScaler, MinMaxScaler
from sklearn.model_selection import GridSearchCV,train_test_split as tts, KFold
from pygam import LinearGAM
from sklearn.pipeline import Pipeline
%matplotlib inline
%config InlineBackend.figure_format = 'retina'
import matplotlib.pyplot as plt
from scipy.optimize import minimize
from matplotlib import pyplot
from nadaraya_watson import NadarayaWatson, NadarayaWatsonCV
Synthetic Data Applications#
rng = np.random.RandomState(123)
# #############################################################################
# Generate sample data
X = 5 * rng.rand(120, 1)
y = 2*np.sin(3*X).ravel()
# Add noise to targets
#y[::4] += 3 * (0.5 - rng.rand(X.shape[0] // 6))
y = y + 0.5*np.random.normal(0,1,len(X))
X_plot = np.linspace(0, 5, 1000)[:, None]
# Fit regression model
train_size = 100
param_grid=[dict(kernel=['polynomial'], degree=np.arange(1, 3)),dict(kernel=['rbf'], gamma=np.logspace(-4, 1, 100))]
# use the sklearn gridsearch
nw_gs = GridSearchCV(NadarayaWatson(), cv=10, param_grid=param_grid)
# use the internal LOO (leave-one-out cross-validation)
nw_cv = NadarayaWatsonCV(param_grid)
# fit k-fold using GridSearch
nw_gs.fit(X[:train_size], y[:train_size])
print("\toptimal bandwidth found: %.2f" % nw_gs.best_estimator_.gamma)
# fit leave-one-out using NadarayaWatsonCV
nw_cv.fit(X[:train_size], y[:train_size])
print("\toptimal bandwidth found: %.3f" % nw_cv.gamma)
# predict
y_gs = nw_gs.predict(X_plot)
y_cv = nw_cv.predict(X_plot)
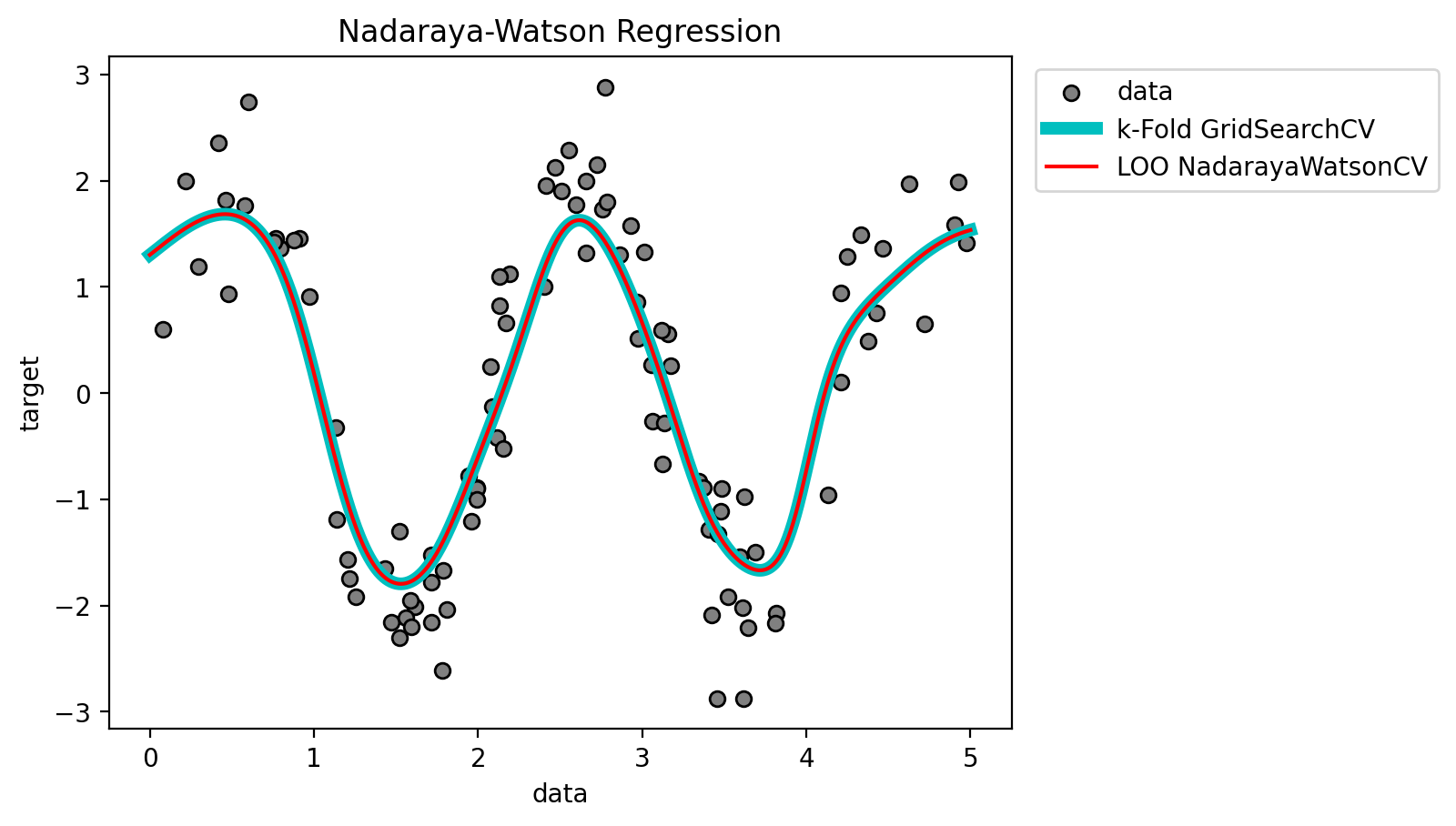
plt.scatter(X[:100], y[:100], c='grey', label='data', zorder=1,
edgecolors=(0, 0, 0))
plt.plot(X_plot, y_gs, 'c', lw=5, label='k-Fold GridSearchCV')
plt.plot(X_plot, y_cv, 'r', lw=1.5, label='LOO NadarayaWatsonCV')
plt.xlabel('data')
plt.ylabel('target')
plt.title('Nadaraya-Watson Regression')
plt.legend(loc = 9, bbox_to_anchor=(1.25,1))
#plt.savefig('fig3.png',bbox_inches='tight',dpi=300)
plt.show()
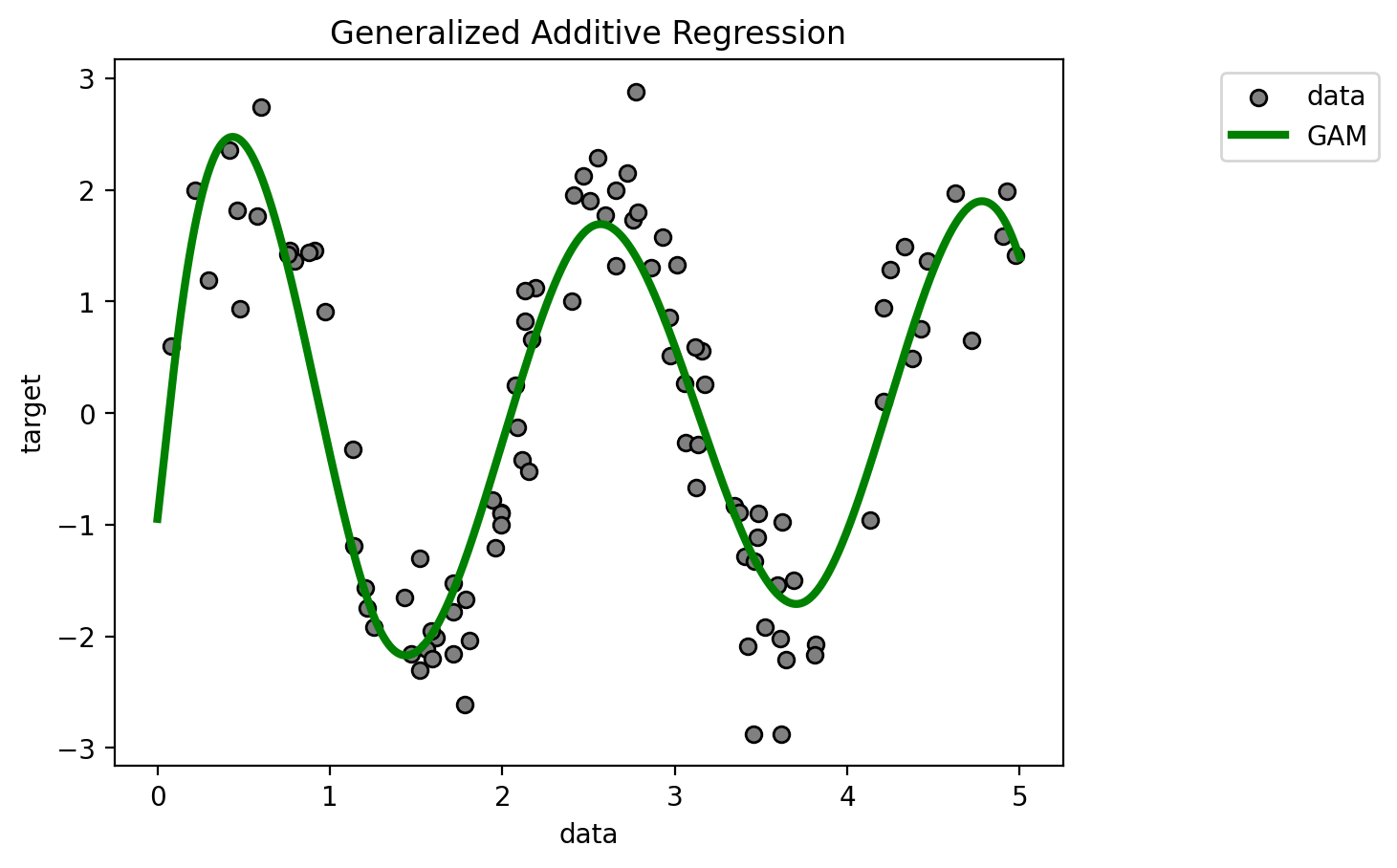
# here you have to guess the "right" number of splines: since we have about 100 data points we should test 6 or 7 splines
gam = LinearGAM(n_splines=ceil(np.log2(len(X[:train_size])))).gridsearch(X[:train_size], y[:train_size],objective='GCV')
100% (11 of 11) |########################| Elapsed Time: 0:00:00 Time: 0:00:00
y_gams = gam.predict(X_plot)
plt.scatter(X[:100], y[:100], c='grey', label='data', zorder=1,
edgecolors=(0, 0, 0))
plt.plot(X_plot, y_gams, 'g', lw=3, label='GAM')
plt.xlabel('data')
plt.ylabel('target')
plt.title('Generalized Additive Regression')
plt.legend(loc = 9, bbox_to_anchor=(1.25,1))
plt.show()
Applications with Real Data#
data_concrete = pd.read_csv('concrete.csv')
x = data_concrete[data_concrete.columns[:-1]]
y = data_concrete[data_concrete.columns[-1]]
ceil(np.log2(len(x)))
11
# compute the number of knots
n_knots = ceil(np.log2(len(x)))
gam = LinearGAM(n_splines=n_knots).gridsearch(x.values, y.values,objective='GCV')
100% (11 of 11) |########################| Elapsed Time: 0:00:04 Time: 0:00:04
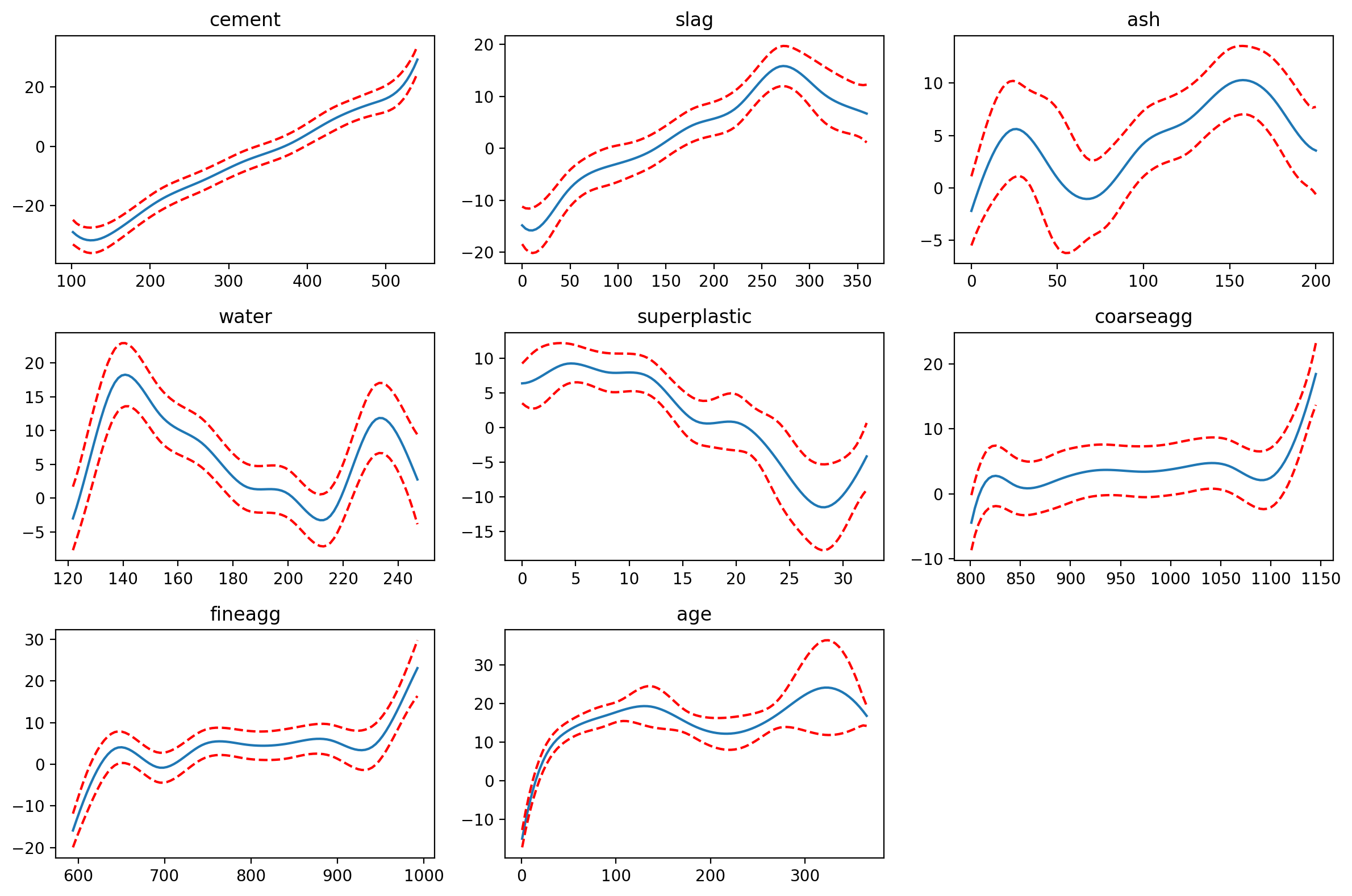
fig = plt.figure(figsize=(8,12))
titles = x.columns
fig.set_figheight(8)
fig.set_figwidth(12)
for i, term in enumerate(gam.terms):
if term.isintercept:
continue
XX = gam.generate_X_grid(term=i)
pdep, confi = gam.partial_dependence(term=i, X=XX, width=0.95)
ax = fig.add_subplot(3, 3, i+1)
ax.plot(XX[:, term.feature], pdep)
ax.plot(XX[:, term.feature], confi, c='r', ls='--')
ax.set_title(titles[i])
fig.tight_layout()
plt.show()
gam.terms
s(0) + s(1) + s(2) + s(3) + s(4) + s(5) + s(6) + s(7) + intercept